MultiRepMacsChIPSeq - Pol2 DeDup
| Home | Overview | Usage | Variations | Examples | Applications | Install |
RNA Polymerase II ChIPSeq Partial Duplication Example
An example of RNA Polymerase II ChIPSeq from mammalian cells. The ChIP samples were prepared with NEBNext Ultra II DNA Library kit and sequenced on a NovaSeq 6000. This ChIPSeq had a total of 105M alignments. After removing 15M optical duplicates (based on a pixel distance of 2500), the UMI duplication rate was 36% resulting in 57M unique alignments. This was one of three comparable biological duplicates.
See the BASH script for the exact pipeline used in processing.
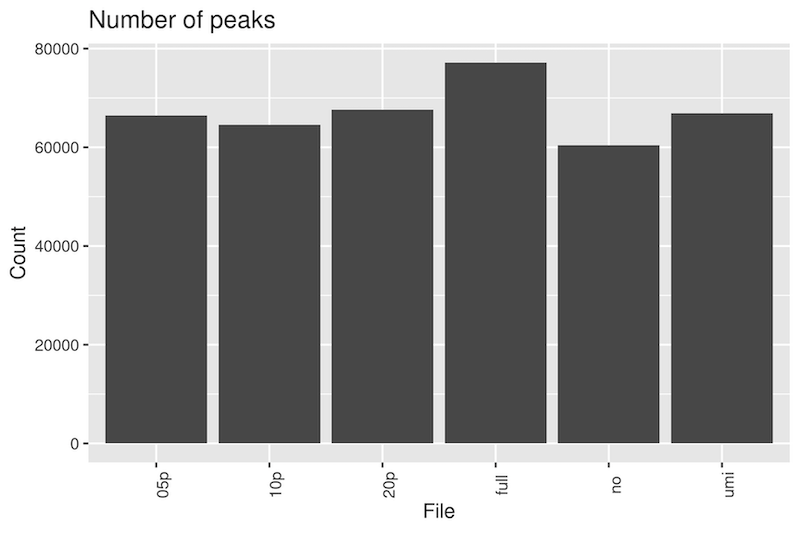
Peak call number
The number of peaks called between different subsets shows more variance than with H3K4me3. In this case, removing all duplicates identifies the least number of peaks, while including all duplicates calls 28% more peaks. The number of peaks called using UMI-deduplicated alignments most closely matches the number called with 5 percent retained duplicates.
The mean peak length was 1140 ± 1883 bp, median 635 bp at a threshold q-value of 2.
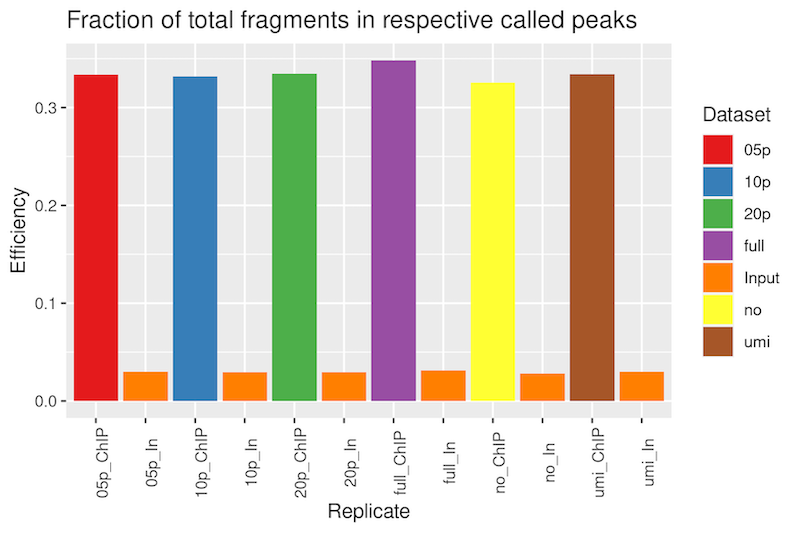
ChIP Efficiency
The fraction of alignments within the respectively called peaks was quite good, with nearly identical fractions between the subsets, ranging from 33-35%.
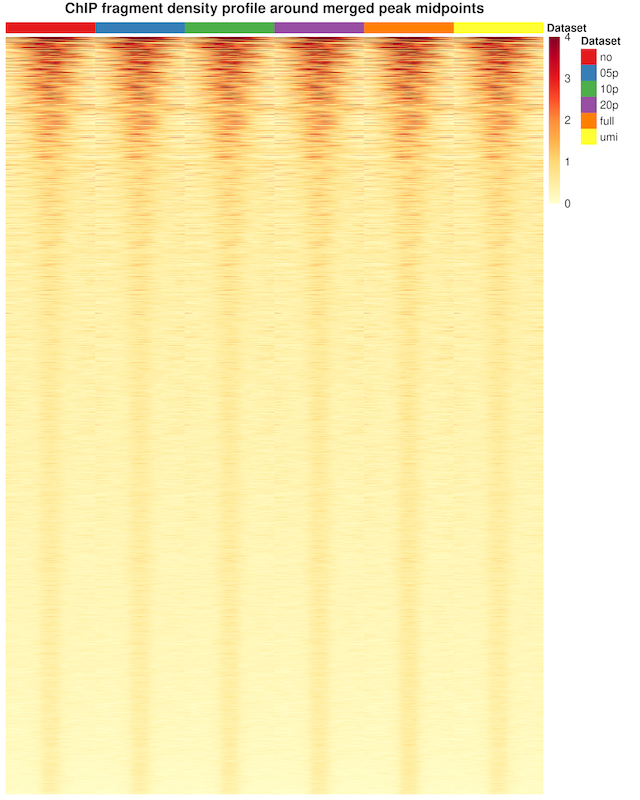
Peak Fragment Coverage Profile
The depth-normalized fragment coverage profile over the called peak midpoint ± 1 Kb shows no difference between the subsets.
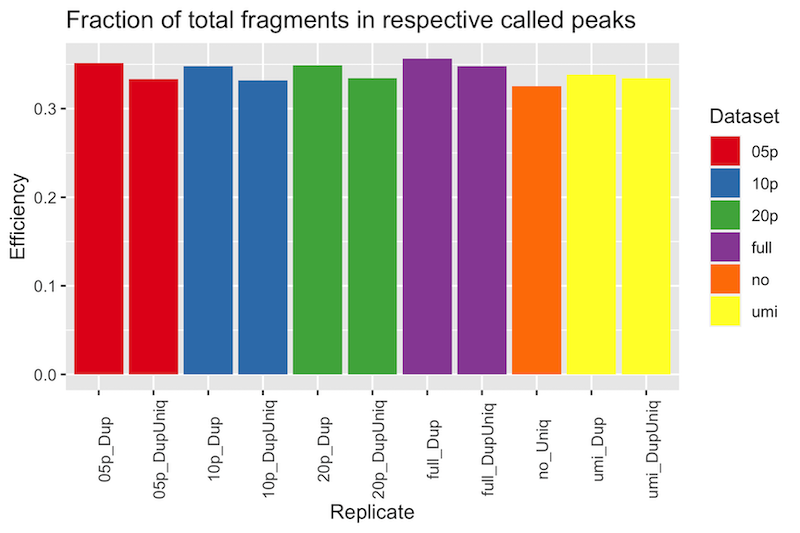
Comparison of Duplicate Alignments Within Peaks
To compare the influence of retained duplicate alignments on the ChIP efficiency, the fraction of alignments (used in the peak call) within each respectively called peaks was compared with the fraction of completely de-duplicated alignments. Each fraction was plotted below.
In all cases, a higher percentage of duplicate-containing alignments were observed in the peaks than completely deduplicated alignments, indicating there are indeed duplicate alignments found within peaks. Notably, however, the difference is only a few percentage points, and more importantly, very little with UMI-deduplicated alignments.
Conclusion
In this RNA Polymerase II ChIPSeq, peak calling with 5 percent retained duplicate alignments most closely matches the true peak call set with UMI deduplicated alignments when considering the number of peaks called. These call sets have 89% spatial overlap (Jaccard statistic; not shown).
When comparing the fraction of alignments with and without duplicates within called peaks, there is very little additional fraction of duplicates within peaks. In fact, the percentages are almost identical with UMI de-duplication, indicating that biological duplication does not in fact play a large contributing factor.